 Image: Janet Iwasa © MIT Press. March 31, 2016
Image: Janet Iwasa © MIT Press. March 31, 2016
|
Programming cells is now a reality, that is a renaisance
of the computational revolution of the past century
Software is opening the door for better hardware
By Juan SIlva @ertwro
Based on Professor Christopher Voigt's lectures on synthethic biology © MIT press[*]
| A group of researchers in the MIT have been developing a programming language based on Verilog, a language to design computer chips, to program the DNA of bacteria. Solving some of the problems of synthetic biology[1]. | ||||||||||||||||||||||||||||||||||||
  Voigt Lab of synthetic biology Voigt Lab of synthetic biology 1/4🎵Is not the words I want to hear from you 🎵
|
  f you think programming is about computers, you probably also think that dancing is about shoes. Or that music is about notes🎵. No, is not like that. Programming is about processes, more generally about life.
The idea of writing a piece of information in a higher language that later gets compiled into a more fundamental jargon, closer to the machine is not new. A set of orders is not something unique to computers. Life does it and has been doing it for a long time (Probably 4.28x106 years).
   One thing one must realize is just how synergic the relationship between language and organism is. Computation underlies everything we see in biology. Is even theorized the brain is a computational adaptation to displacement. Computation is natural and has always been self-reinforcing.
|
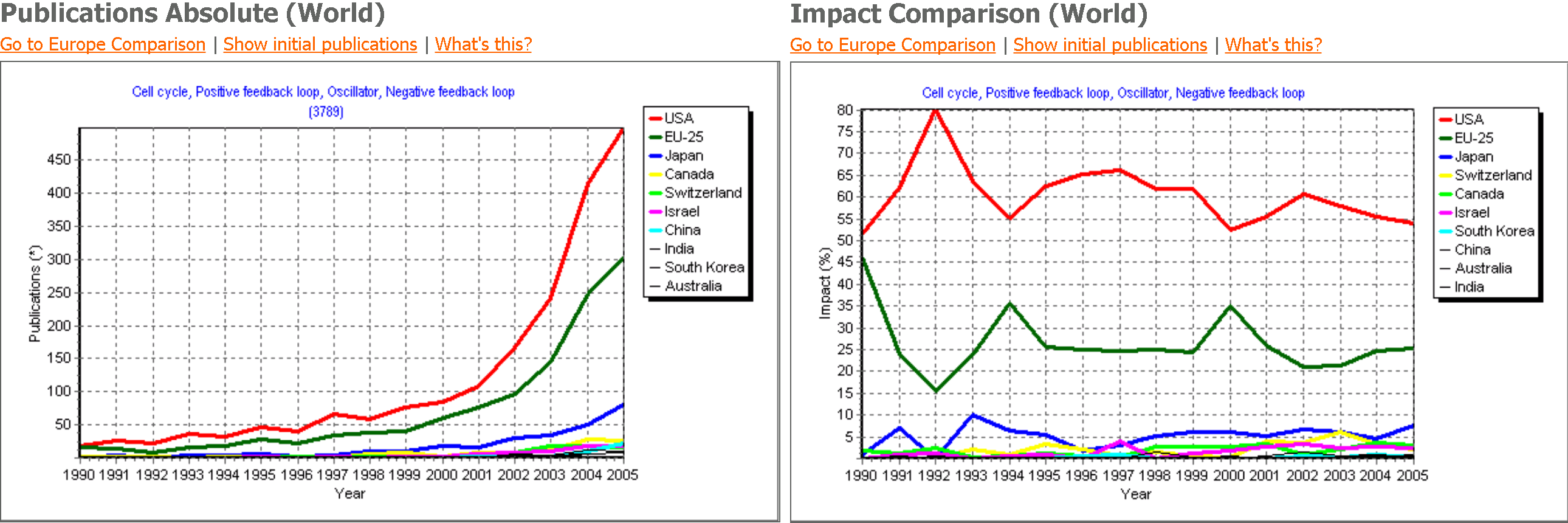
 The charts show the temporal evolution of the publications in the field
The charts show the temporal evolution of the publications in the field|
Bioengineering is already a reality in its infancy. At the moment 2% of all the American economy depends on products derived from engineered biology.[2] Genetically modified cells that give us products like Medicines. foods, plastics, etc.
What all those products have in common is the fact that their chemical structure is quite simple. No more than a few carbon, hydrogen and oxygen atoms. The necessary number of changes it requires is quite small. Just a couple of genes.
   The potential to mass produce chemically complex products has a type of sophistication that current laboratory techniques can only dream of. Cells and biomolecules are nanomachines and harnessing their power could mean a materials revolution no short of what the industrial revolution did. This requires hundreds or thousands of modifications. Not only that but coordinating when they go On or Off. Changing mistakes in the midst of the machine functioning.
 |
2/4🎵 More than words🎵
🎵 Is all you have to do to make it real ...🎵
  ells use a huge regulatory network. DNA, proteins, and ARN are interacting and this can be seen as computations[3]. In the 70's people started referring to this arrays as switches or logic gates. Borrowing from the language of electrical engineering. In the case of Bacterias like E. coli, the networks get significantly big[4].
|
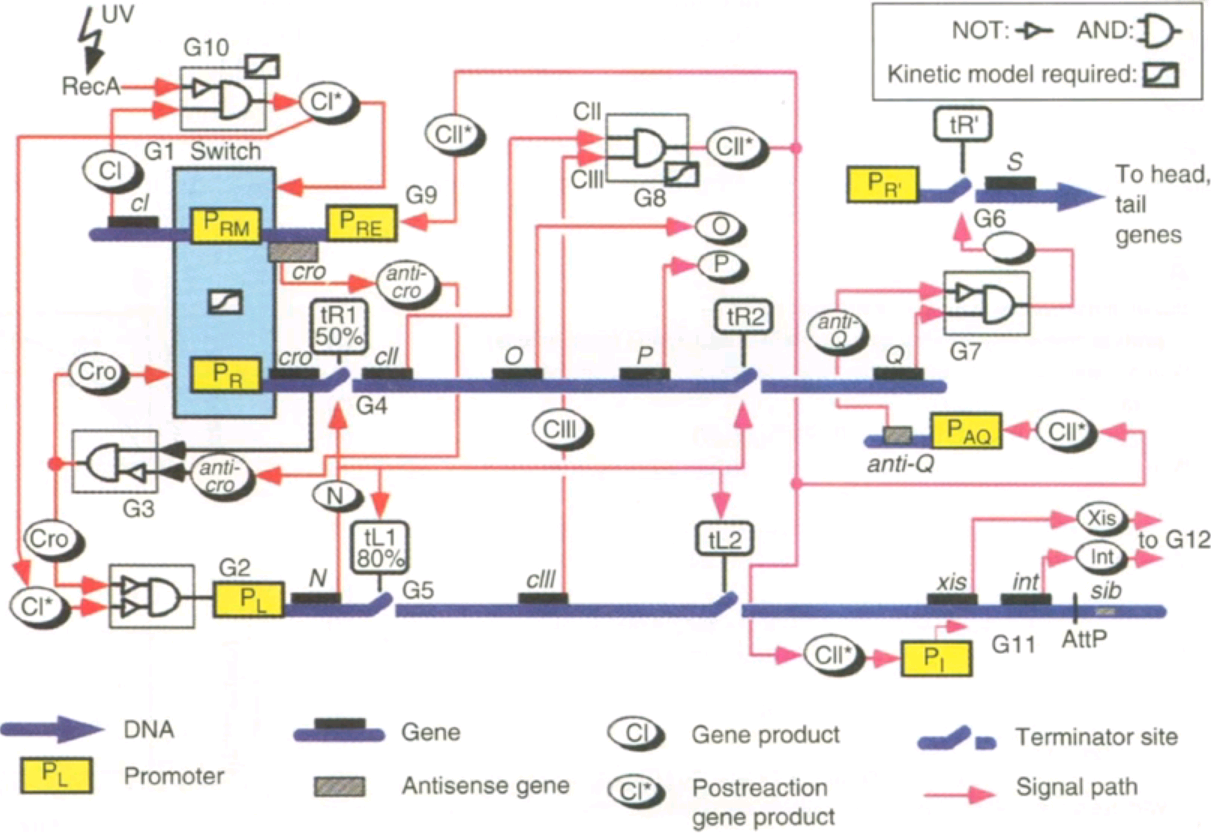
 10.1126/science.7624793 Fig, 4. Regulatory pathway of a virus after it enters a cell. It needs to decide if
10.1126/science.7624793 Fig, 4. Regulatory pathway of a virus after it enters a cell. It needs to decide if kill the bacteria and spread or integrate into the genome to stay in existence for a longer time.
|
As with most of the developments in biology, it has a huge surface of trial and error to advance a single step. For perspective, while DNA stores information in pairs of bases and one would expect 2bits of information per combination is not so. In reality, it only stores 1.8bits per nucleotide due self interference errors.
Although since it has 4[5] building blocks it could create at least two orders of magnitude more space than what already holds. Currently is the single most dense form of information storage at a whopping 1.28 PetaBytes/gram and a possible 215 PetaBytes/gram. To give you an idea, this means all the information ever created by humanity could be stored in no more than 10 cubic meters or a couple of pickup trucks.[6]
|
|
|
As opposed to designing by moving individual atoms, which has the most known potential with quantum effects that are super resource expensive, biology offers huge potential scalability. Is not cheap as writing and reading information is at $20k per PetaByte.
The problem to scale this comes in many forms. One of them is design. Since most of the advancements have been achieved by trial and error there's not enough framework to determine how they are gonna interact when they are combined. So software doesn't really exist to aid in that process. The current architecture is complex, fragile and expensive to write. |

3/4🎵 What would you do🎵
🎵 If my heart was torn in two?🎵

 he state of the art in genetic artificial computation at this point is closer to the beginnings of machine computation, I could say we are in the 1960's of biologic computing. First, the hardware that performed basic operation was build. Then the apparition of the microchip and from then on an explosion in engineering and software design.[7]
   Every logically complete operation can be completed with NOT
(¬) & AND(^) as conectives and the combination of both is ↓ NOR ¬(p v q) gates. Is a function that receives two inputs and gives one output  . Every ordinary computational system can be built with it. In fact, back in the 50's the apollo program was completed by using a set of coumputers that were constructed using functionally complete singletons[8]. . Every ordinary computational system can be built with it. In fact, back in the 50's the apollo program was completed by using a set of coumputers that were constructed using functionally complete singletons[8].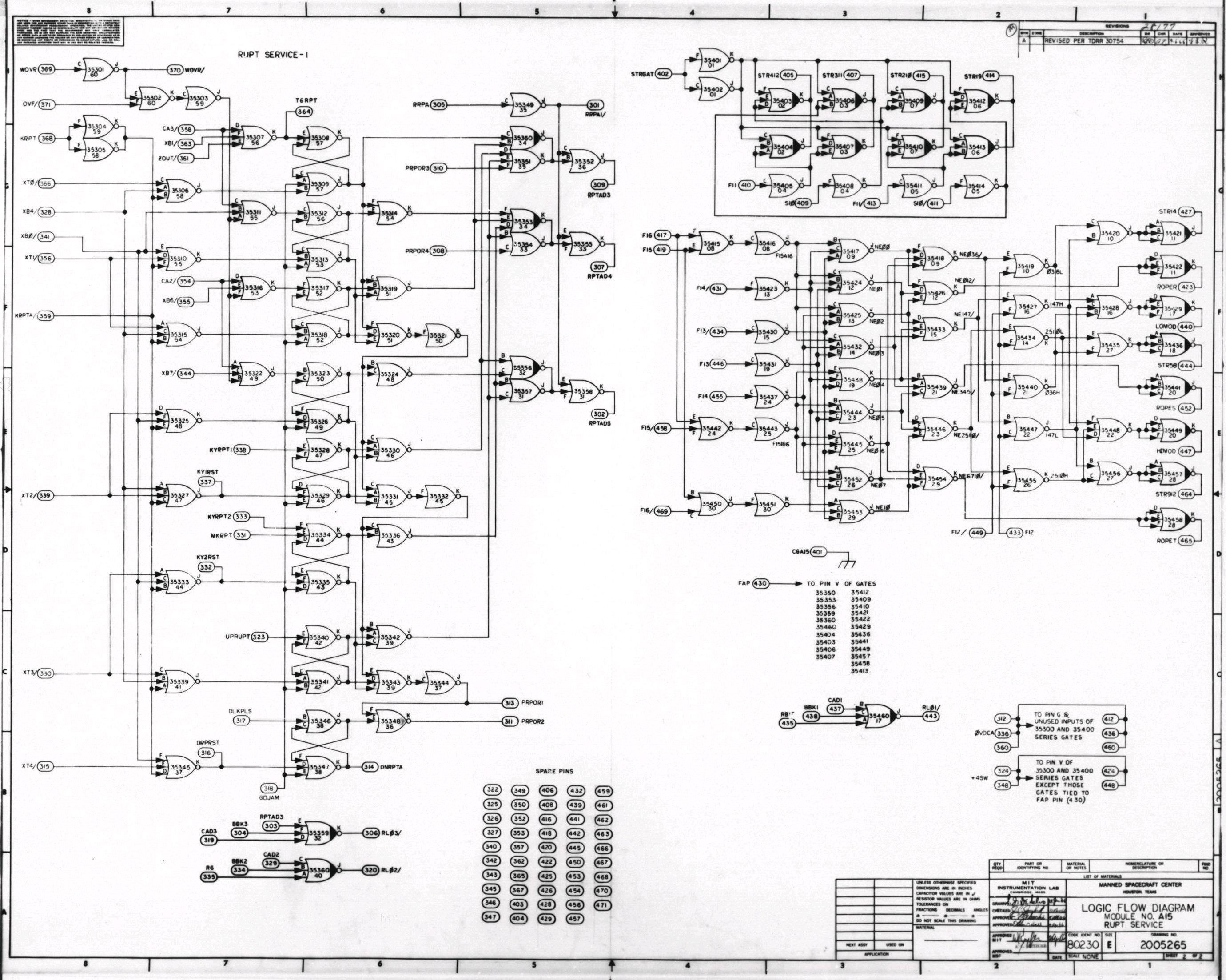  5600 interconected triple input NOR gates 5600 interconected triple input NOR gatesApollo Mission schematics, guidance systems. |
 In the case of cells, inside the nuclei, a sensor is a piece of DNA that allows a cell to respond to a signal and thus control the expression of a gene. When the signal interacts with the sensor
A) it joins at the promoter. This causes a high flux of RNA polymerase (RNAP). An in the absence of this interaction the flux of RNAP is slowB)
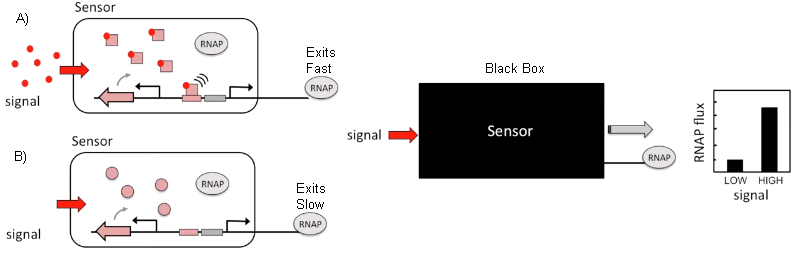
Like this disregarding the content it can be seen as a black box and just compute the difference in flux to treat it as a circuit.[9] In the case of it as a circuit, it has RNAP as an input and as an output. Due to this, you can interconnect the circuits in a universal Turing machine-like fashion. In this case the function of silencing promoters by using repressors has been used as a NOT gate 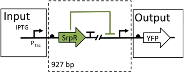 . With two repressors in line, the same effect of a NOR gate can be achieved. The NOR gate is the Heart of this solution. . With two repressors in line, the same effect of a NOR gate can be achieved. The NOR gate is the Heart of this solution.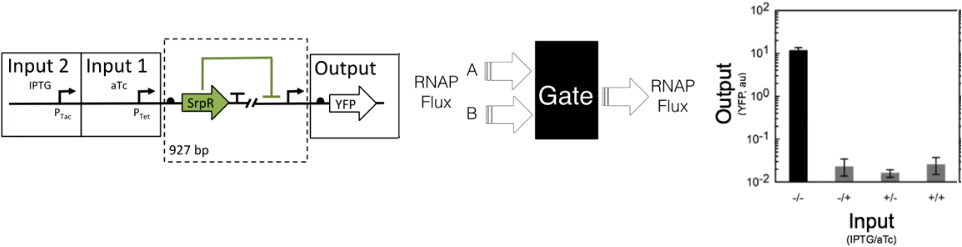
|
|
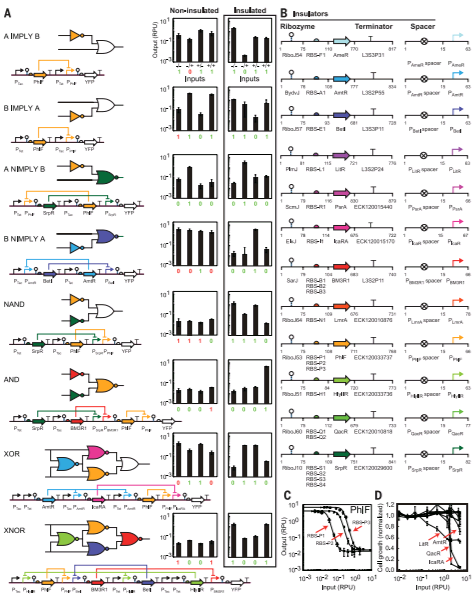
In order to select the appropriate language borrowed from electrical engineering, researchers selected Verilog. A type of hardware independent language where people can create a function that can be compiled into DNA. Lower order languages that are closer to the machine are still used by chip designers today. The web program is called Cello. 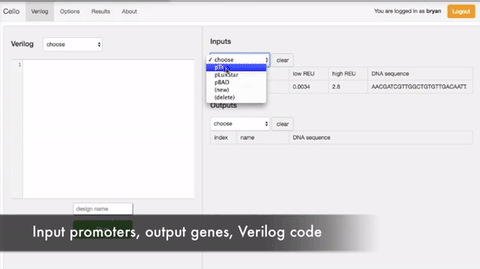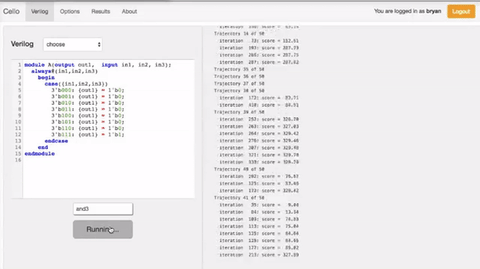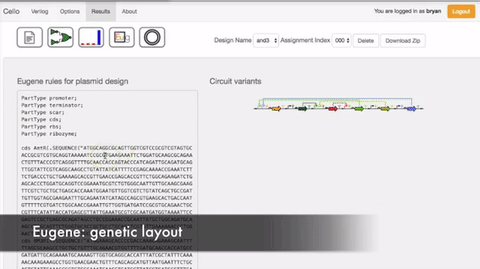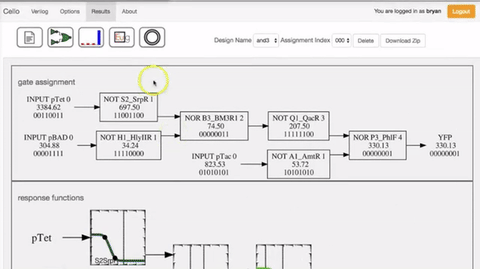 The user writes the logic functions using Verilog commands. Once the user runs the compilation the program automatically finds the necessary diagram and gates configuration form the library of insulated gates.
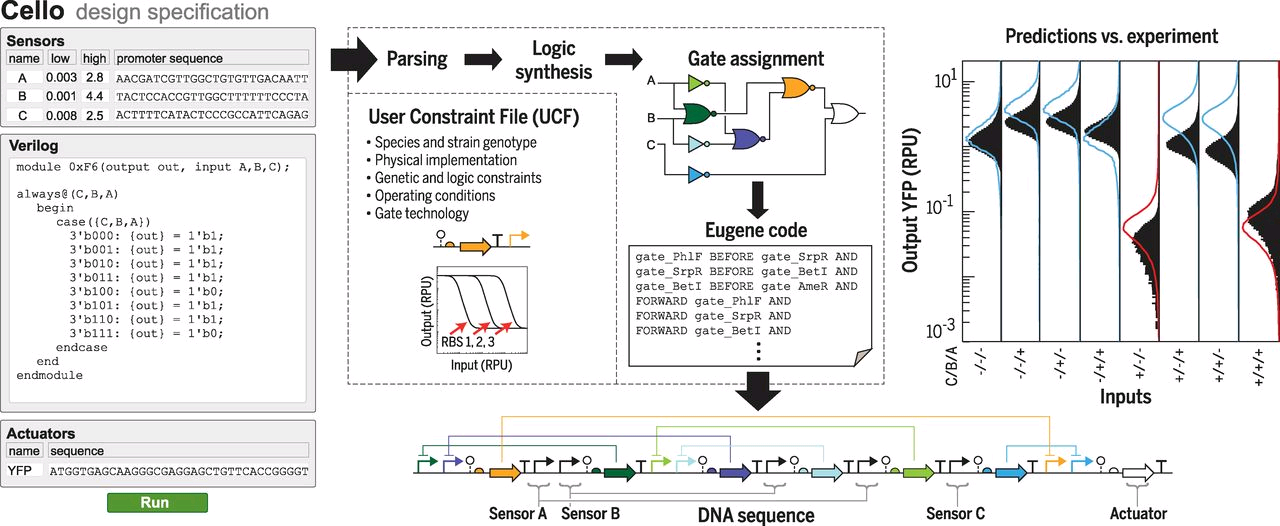  Figue: Shows the order of the sequence of comands Figue: Shows the order of the sequence of comands |
|
Due to the spatial orientation at the interior of the cell, interference between repressors could happen. There have been identified 16 different types or non-interfering repressors that can be used to construct gates. Is also necessary a form of insulation that can be used in multiple configurations in what's called the Gate library[10].
Once the code is compiled, the output of the program is a DNA sequence ready to be tested in vivo
|

4/4🎵 What wold you say🎵
🎵 If I took those words away?🎵

 ack in 2013, the Reproducibility Project was launched by Amgen. Researchers tried to replicate the experimental results of 53 lanmark cancer studies. Later to find that 47/53 couldn't be replicated. This was a huge blow for pre-clinical research standards.[11]
Is probably not a mystery how this happened. Is not necessarily that all the studies were lies or the original researchers cheated. Is probably that the registry of everyday data and protocols are unreliable by nature. If you ever check a researcher's Lab Notebook you will find that is close to an alchemist book. Filled with notes that don't make sense at the moment for use by other people. Even the most well kept will take for granted many things that could be vital for the research.
  Github read me. Contains comparissons Github read me. Contains comparissonsUsing Javascript Something similar is said about code. "Nobody can use other person's code, you shall use your own". Still, all civilization is a construction over the work of others. What happens is that the creation of standards requires interdisciplinary support to neophytes and experts in fields their own and outside their fields.
Is a huge effort but is necessary. This is an example of how proper changing translation helps people:[12]
science requires imagination and hard work. Words to guide others that will come. Applied to problems new an outside our domain of expertise. Every new endeavor is a reinvention that creates translation knowledge that allows hardware improvements for everyone.
   🎵 I ever needed you to show 🎵  🎵 That you love me 🎵 🎵'cause I'd already know🎵 REFERENCES: [1] Kwok, R. (2010). Five hard truths for synthetic biology. Nature, 463(7279), 288–290. [6] Bohannon, J. (2011). The Life Hacker. Science, 333(6047), 1236–1237. |