Introduction
Ever since our emergence as a species, humans have suffered from disease. Amongst the diseases endangering humanity, those caused by pathogens that represent nearly every branch on the tree of life, have represented and continue to be a potent and direct biotic threat to our survival. Consequently, there has been lots of interest in the agents that cause disease, their biology and the means by which disease can be prevented or eradicated. Interestingly however, the pathogens that affect plants form an indirect but as of yet greater biotic threat to our survival. In this review, I will therefore describe our understanding of Plant-pathogen interactions to discuss the factors that determine pathogenicity. Specifically, I will describe the current concepts that are thought to underpin Plant-Microbe interactions and form some of the reasons as to why some microbes are in fact, pathogenic and affect our food crops.

Resistant (L) and susceptible (R) plant species inoculated with Phytophthora. Tissue collapse and lesion growth is characteristic of susceptibility (right leaf) whereas resistant plans appear unaffected (Left leaf).
Studies of pathogens and their biology
Pathogens have been known to affect plants for centuries with early written records describing the occurrence of disease and famine. Ever since then, there has been much interest in the agents that cause disease as well as a focus on how outbreaks on crops can be prevented. This led to the emergence of the Plant Pathology & epidemiology fields, seeking to identify and understand the (population) biology of pathogens. In addition, geneticists have long sought to define the factors specifying disease and resistance on crops. Consequently, a vast number of plant pathogens, collectively affecting every single plant or crop, have been identified and described. In addition, information on the genetic factors, present in plants, that affect disease outcomes were initially described along with some mechanistic insights. With the emergence of genetics, molecular biology, genomics, computational biology and functional genomics however, the understanding of pathogen biology has dramatically improved. The observation that pathogens are often specialists on a limited set of hosts, has prompted much interest in understanding the basis of host susceptibility and resistance (immunity) (Figure 1).
The Molecular basis of Plant-Pathogen interactions
Plants are continuously bombarded by a diverse array of microbes that can cause disease. In most cases, infection is limited through the perception of Pathogen (or Microbe) Associated Molecular Patterns (PAMPs or MAMPs) by Pattern Recognition Receptors (PRRs) (Abramovitch et al., 2006, Brunner et al., 2002). Recognition results in PAMP triggered immunity (PTI) and features a marked shift in transcriptional activity towards defence (Figure 2). In a select few cases however, pathogens successfully infect plants of a given species. This suggests that host immune responses are suppressed or evaded, a pathogen characteristic that suggests an evolutionary basis for specialization (Jones and Dangl, 2006). Genome sequencing projects, combined with the development of computational pipelines and high throughput functional assays, have led to the identification of factors responsible for pathogen virulence (or pathogenicity) and therefore has revolutionized our thinking about plant pathogens (Oliva et al., 2010, Kamoun, 2003, Kamoun, 2006, Kamoun, 2007). State of the art models emanating from the field now describe pathogen molecules that are secreted and delivered into host tissues (effectors) to subvert host immunity (Figure 2).
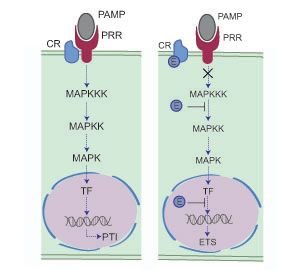
Mechanistic model of plant-microbe interactions. Left, a microbe is recognised by a plant PRR, leading to a successful defense response. In a susceptible interaction (right panel), pathogen effectors disturb plant responses and compromise the immune response. this allows the pathogen to infect and cause disease. Figure modified from Howden & Huitema (2012).
Pathogens topple the host immune signaling network
If plants employ vast signaling networks to regulate immune responses and fend off most microbes, pathogens must be able to topple such systems to effect disease. Although this notion has been widely accepted for some time, the mechanisms by which immune suppression occurs, has remained elusive until recently. Both prokaryote and filamentous (eukaryote) pathogens form specialized structures that interface with the host. These structures are the sites where subsets of effectors (cytoplasmic effectors) are translocated into the host cell where they carry out their intended functions (Whisson et al., 2007). Systematic studies have since implicated effector repertoires that disrupt the immune signaling network and effect disease (effector triggered susceptibility or ETS) (Mukhtar et al., 2011). Effector interaction studies have identified host targets in plants, providing functional information on both effector activity and the role of their intended plant target towards immunity. Although the targets and their roles have now have been defined, exact mechanistic detail has been lacking. It has however lent significant support towards a model that describes the induction of susceptibility by a given pathogen (effector triggered susceptibility or ETS).
Pathogen effectors trigger susceptibility in their host(s)
Only recently, significant progress has been made towards improving our understanding of effector mode of action towards ETS. It is generally assumed that the interaction between an effector and its host target, results in a modification that subsequently changes its cellular fate and triggers susceptibility (Kamoun and Goodwin, 2007). This modification is achievable through two general mechanisms. The first mechanistic model presumes that the effector (and its functional domain) is directly responsible for the target modification event. A direct interaction combined with effector enzymatic activity, ensures modification of the target. An alternative mechanism relies on the host machinery to modify the effector target. In this model, the effector binds its target and signals modification or re-localisation by the host cell. Evidence supporting both mechanistic models has recently been reported (Stam et al., 2014) but it is likely that other mechanisms will be unveiled in the future. Regardless, effector-mediated target modifications underpin changes in the activity or cellular fate of important immune modulators and are critical for ETS or disease establishment.
Pathogen host specialization and evolution
Effectors are the principal determinants of virulence or pathogenicity. One major unknown however, is why all members of a plant species can be resistant to a given pathogen species, whereas others are susceptible. The molecular basis of this resistance on the species level (termed non-host resistance or NHR) remains elusive, but two major effector based hypotheses explaining this phenomenon have emerged (Schulze-Lefert and Panstruga). The first hypothesis assumes that in closely related plant species, pathogen effectors can interact with their host targets and in principle, topple the immune signalling network. Nonhost plants in this scenario however, carry one or more receptors that recognise either multiple or essential effectors that are conserved in a pathogen species which then leads to (non-host) ETI (Schulze-Lefert and Panstruga). The second model describes a situation where two plant species are distant relatives (e.g. Tomato vs Arabidopsis). The nonhost plant carries an immune signaling network whose components cannot be modified by the pathogen. In this model, co-evolution between host and pathogen and divergence between host and non-host, has resulted in a loss of specificity towards key signaling components in non-host plants (Schulze-Lefert and Panstruga). Consequently, the inability to suppress PTI leads to avirulence for the pathogen. Critically, these models are not mutually exclusive and are likely to be substantiated in the future.
Conclusions
Host-pathogen interactions are complex associations whose outcomes are determined by the interplay of both host and pathogen encoded factors. With both secreted pathogen proteins (effectors) and host immune signalling components present in plants during infection, it is clear that the plant cell forms a molecular battleground where immunity and susceptibility to pathogens is specified. Therefore, defining the basis of pathogenicity will lead to a multitude of distinct answers, each equally applicable and valid with regards to the original question. The observations that (i) pathogens have evolved the means to cause disease and (ii) hosts maintain a large and complex signaling network, implies the involvement of great evolutionary forces, ultimately specifying host-microbe relationships. Genome wide comparisons between (non) pathogens and their (non) hosts will expand our comprehensive view on pathogenesis further, mitigate crop loss and the indirect threat to our survival.
REFERENCES
ABRAMOVITCH, R. B., ANDERSON, J. C. & MARTIN, G. B. 2006. Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol, 7, 601-11.
BRUNNER, F., ROSAHL, S., LEE, J., RUDD, J. J., GEILER, C., KAUPPINEN, S., RASMUSSEN, G., SCHEEL, D. & NURNBERGER, T. 2002. Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. Embo J., 21, 6681-6688.
JONES, J. D. & DANGL, J. L. 2006. The plant immune system. Nature, 444, 323-329.
HOWDEN, A. J. M. and HUITEMA, E. (2012). Effector-triggered post-translational modifications and their role in suppression of plant immunity. Front. Plant Sci. 3:160 10.3389/fpls.2012.00160
KAMOUN, S. 2003. Molecular genetics of pathogenic oomycetes. Eukaryotic Cell, 2, 191-199.
KAMOUN, S. 2006. A Catalogue of the Effector Secretome of Plant Pathogenic Oomycetes. Annu. Rev. Phytopathol., 44, 41-60. KAMOUN, S. 2007. Groovy times: filamentous pathogen effectors revealed. Curr Opin Plant Biol, 10, 358-365.
KAMOUN, S. & GOODWIN, S. B. 2007. Fungal and oomycete genes galore. New Phytol., 174, 713-717.
MUKHTAR, M. S., CARVUNIS, A. R., DREZE, M., EPPLE, P., STEINBRENNER, J., MOORE, J., TASAN, M., GALLI, M., HAO, T., NISHIMURA, M. T., PEVZNER, S. J., DONOVAN, S. E., GHAMSARI, L., SANTHANAM, B., ROMERO, V., POULIN, M. M., GEBREAB, F., GUTIERREZ, B. J., TAM, S., MONACHELLO, D., BOXEM, M., HARBORT, C. J., MCDONALD, N., GAI, L., CHEN, H., HE, Y., VANDENHAUTE, J., ROTH, F. P., HILL, D. E., ECKER, J. R., VIDAL, M., BEYNON, J., BRAUN, P. & DANGL, J. L. 2011. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science, 333, 596-601.
OLIVA, R., WIN, J., RAFFAELE, S., BOUTEMY, L., BOZKURT, T. O., CHAPARRO-GARCIA, A., SEGRETIN, M. E., STAM, R., SCHORNACK, S., CANO, L. M., VAN DAMME, M., HUITEMA, E., THINES, M., BANFIELD, M. J. & KAMOUN, S. 2010. Recent developments in effector biology of filamentous plant pathogens. Cell Microbiol.
SCHULZE-LEFERT, P. & PANSTRUGA, R. A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation.
STAM, R., MANTELIN, S., MCLELLAN, H. & THILLIEZ, G. 2014. The role of effectors in nonhost resistance to filamentous plant pathogens. Frontiers in Plant Science, 5, 582.
WHISSON, S. C., BOEVINK, P. C., MOLELEKI, L., AVROVA, A. O., MORALES, J. G., GILROY, E. M., ARMSTRONG, M. R., GROUFFAUD, S., VAN WEST, P., CHAPMAN, S., HEIN, I., TOTH, I. K., PRITCHARD, L. & BIRCH, P. R. 2007. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115-118.
Original source for Figure 2:
Original Source: