To sequence your entire genome costs under $1,000 today; what you get is the precise order of the 3 billion nucleotide base-pairs (adenine-thymine, cytosine-guanine) in your DNA.
But you don't need full genome sequencing to get a good grasp of your genetics.
You could use services like Ancestry, 23andme, and the like to check only for certain positions in your genome (~1 million for 23andme - as of 2016). The service costs ~$100 and the positions they check for have shown to be associated with different physiologic/psychologic conditions (either positive or negative).
When 23andme checks your genome at these positions, they compare the nucleotide at one position in your genome to the nucleotide in a reference genome. It they do not match, there is a mutation in your genome. What they do is actually to look for SNPs (single nucleotide polymorphisms). According to the NIH Handbook of Genetics:
"Each SNP represents a difference in a single DNA building block, called a nucleotide. For example, a SNP may replace the nucleotide cytosine (C) with the nucleotide thymine (T) in a certain stretch of DNA."
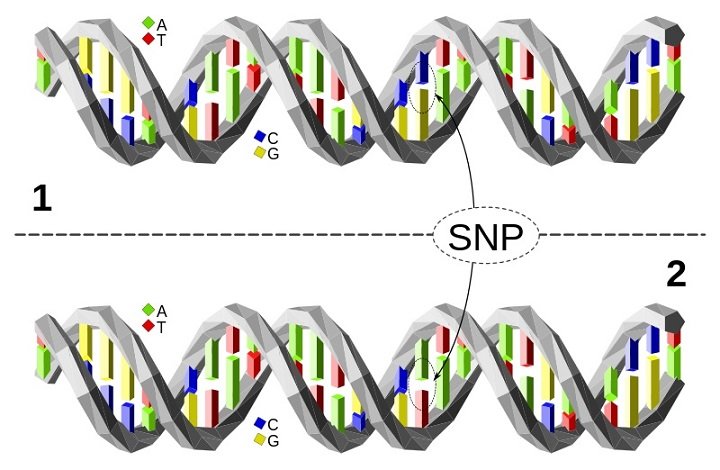
DNA replication/maintenance/repair is not bulletproof; errors can occur.
Thus, we normally have ~10 million SNPs in our genomes. Since some of these mutations occur in non-protein-coding regions they may not be associated with any pathophysiologic conditions. Those occurring in coding or impactful regions may be more involved in disease. As I wrote previously:
"While many of these SNPs are not determinative by themselves (in isolation), there are various that can heavily impact or characterize a disease state, like the cystic fibrosis – CFTR – associated SNP, the BRCA1 SNP associated with higher risk for developing cancer, the MSH2 SNP associated with Lynch Syndrome and with colorectal cancer, and so on."
My purpose with this post is to introduce some SNPs associated with obesity. Those carrying these SNPs in their genomes may be more susceptible to developing obesity and type 2 diabetes.
Genetic Mutations (SNPs) associated with Obesity
1. APOA2 - gene that codes for Apolipoprotein A-II
- involved in lipid metabolism
- defects in APOA2 => APOAII deficiency and higher cholesterol levels [source].
Single Nucleotide Polymorphisms of APOA2 (currently available):
- rs5082
The risk genotype for this position is CC, meaning that if you have CC in your DNA at this position, you could be at higher risk for increased plasma lipids compared to people carrying the normal genotype. Here's the study that found this association: study.
Read more about rs5082.
2. APOA5 - gene that codes for Apolipoprotein A-V
- component of HDL
- involved in triglyceride control
- mutations/defects in APOA5 => higher triglycerides, higher lipoprotein levels [source].
Some of the Single Nucleotide Polymorphisms of APOA5 (currently available):
– rs2075291
The normal genotype (the nucleotides in the reference genome at this position) is CC. People from Asian descent who carry AA or AC at this position are at 4 times higher risk for abnormal triglycerides. In this case, it would make sense avoiding a high fat diet. See the study that found this association: here.
Read more about rs2075291.
– rs662799
The normal genotype (from the reference genome) at this position is AA. People carrying AG or GG at this position are up to 2 times at higher risk of suffering a heart attack, despite the fact that they are less susceptible to gain weight through a high-fat diet. Two studies associated with this SNP are: here and here.
Read more about rs662799.
Ending Thoughts
To reiterate, you can use services like 23andme or Ancestry to determine the SNPs you may carry. I am more familiar with 23andme as I have consulted people who sequenced their genomes with this service. The raw data (as text) you get from them looks something like this:
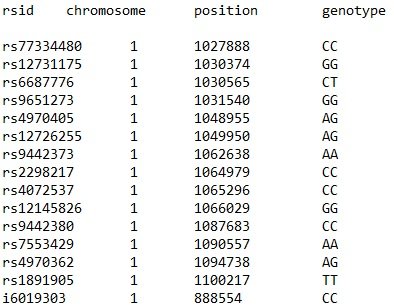
Now, you could do the investigation yourself: take each SNP and browse through SNPedia and medical databases to see associations with this genetic mutation. But, going through 1 million SNPs (at the moment) is an unattainable goal, especially if most of these SNPs may not have major impact on your overall health.
You could use paid services like Promethease that does it automatically: takes your raw data, does the work, and 5 minutes later it returns a report with tables containing each of your SNP, the reference genome position, and the conditions associated with the SNP, as well as links to studies that are relevant to that SNP. You could then sort the SNPs based on the level of associated risk they carry. And this is a much more feasible approach, should you decide to know more about your genome.
Genetics play a big role in our health, but lifestyle factors and epigenetics carry heavy roles as well. In some cases it may be important to know your genetics, while in some others it may be not. It's up to you to decide.
In the second part I will continue my discussion on SNPs associated with obesity.
To stay in touch with me, follow @cristi
Credits for Image: [David Eccles CC BY 4.0 via Wikimedia Commons.
Cristi Vlad, Self-Experimenter and Author
