In the 19th century, Charles Darwin published his theory of biological evolution in "On the origin of species". According to his research, all species of organisms live under the priniciple of natural selection. This concept is shaped from the difference in the ability of the individual to survive, reproduce and compete. The reason for that is, by reproducing, a certain, herited, genetic variation occurs, which affects the phenotype of the individual. As there is a competition between the individuals of a species for ressources like food, sexual partners and living space, surviving is determined by who's better adapted.

Charles Darwin ( 12 February 1809 – 19 April 1882 )
This competition is described as the "struggle for live". As better adaptions aren't the result of an inner need from the individual like Lamarks theory suggest, but from the recombination of genetic information, randomly better adapted individuals have a higher chance to survive. "Survival of the fittest" ist the term describing this process. Taking a giraffe in the steppe as an example, a slightly longer neck was often already an advantage big enough to safe the survival of the animal because it could reach food which the smaller animals couldn't. Over a long period of time, percentagewise far more giraffes with a short neck will die. The bigger giraffes reproduce and have a higher chance of getting descendants with a longer neck due to their DNA. Darwin hadn't the information about DNA which we have today. Phylogenetic deals with the relationship of different species.
How can we determine if two animals are related?
The comparing anatomy did this by looking at homolgies. Comparing two animals e.g. shark and human, we can for example find out that the structure of a shark's scale is in it's buildup very similar to a human tooth. Rows are also used as they are part of the criteria to determine if there is a homology. Similarities play an important role in the first test too. They try to find the relationship on a smaller level:
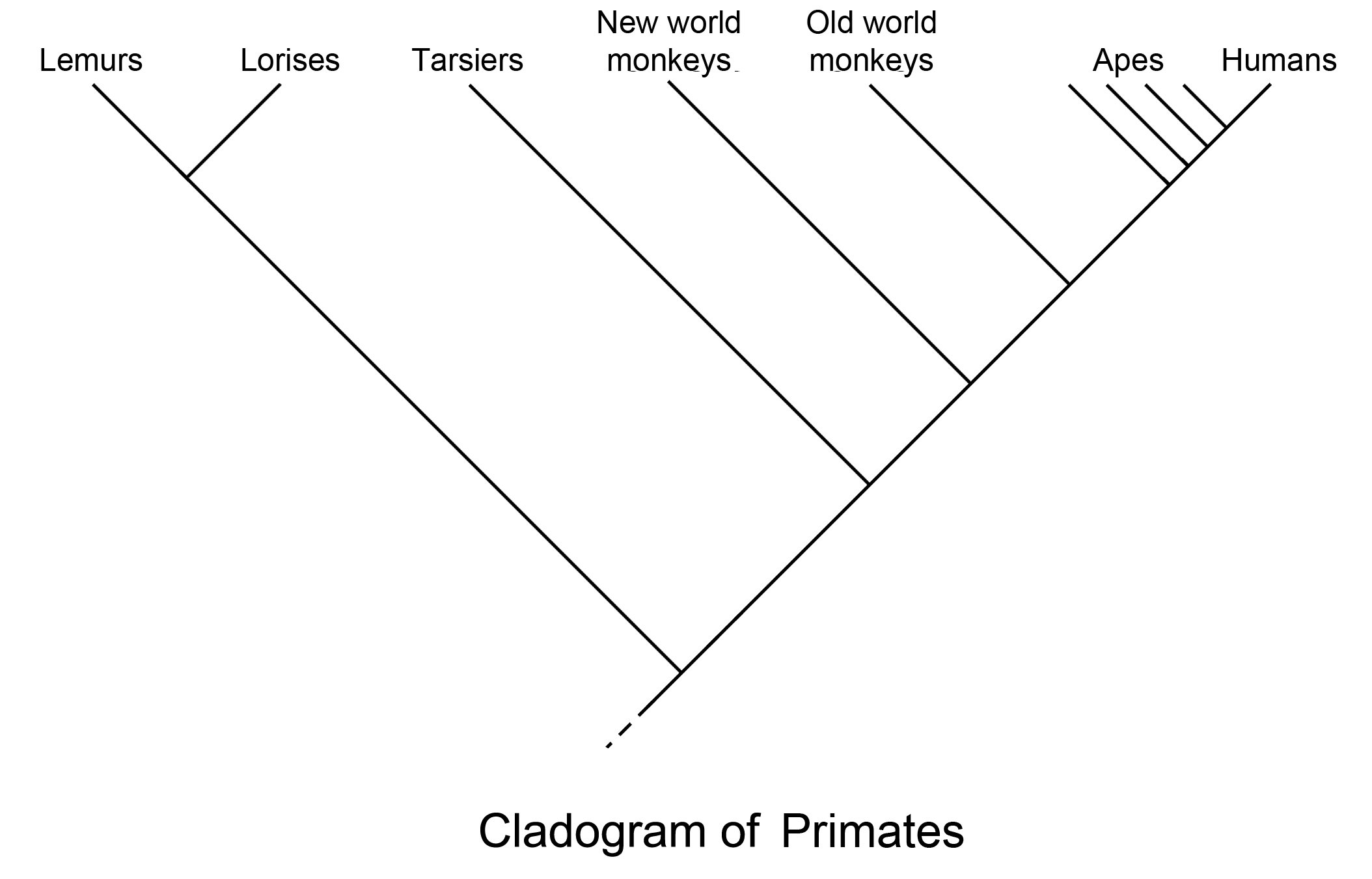
Phylogenetic root tree
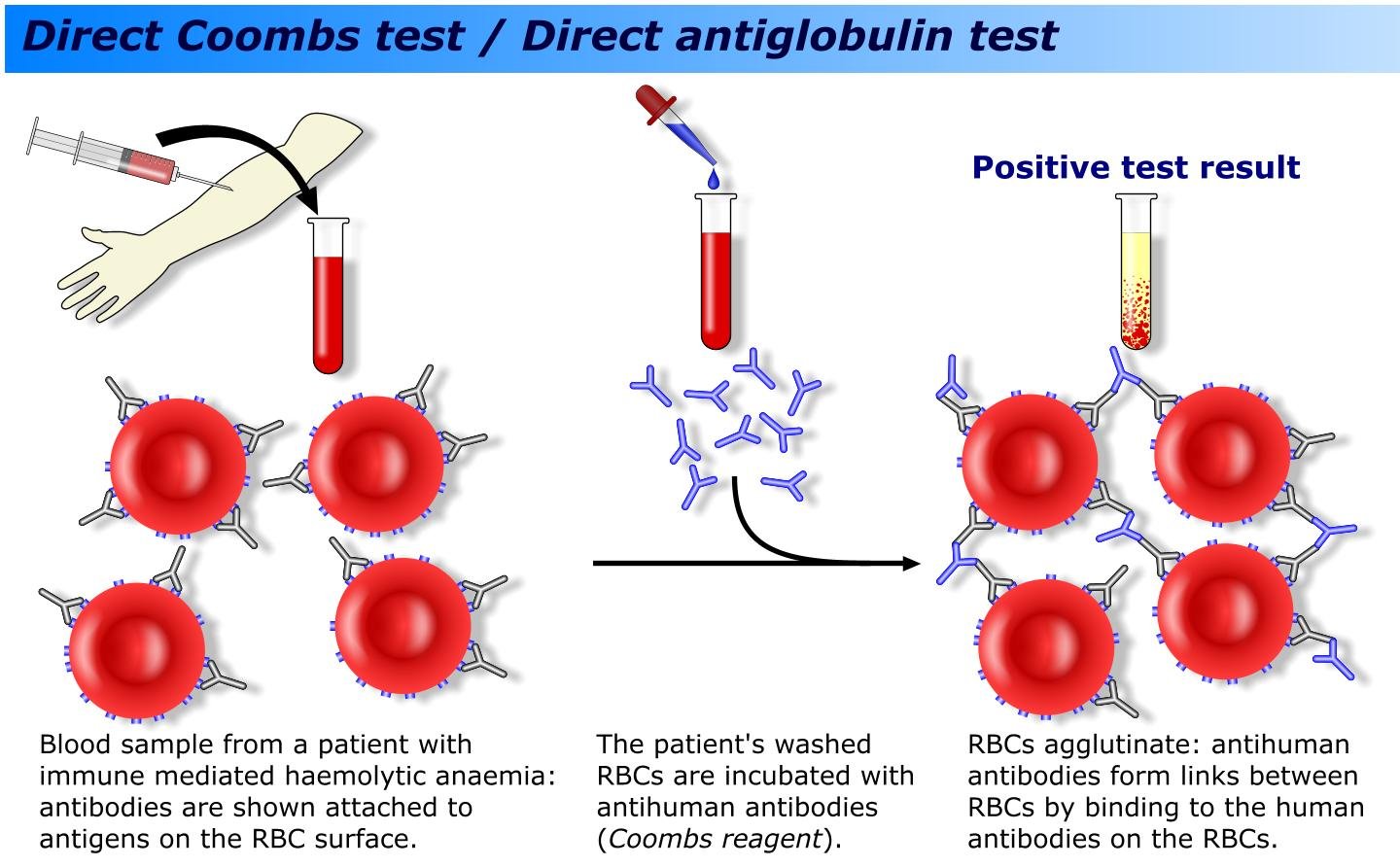
Precipitin test
Human blood serum get's injected into an animal e.g. a rabbit. After some time it's blood wird removed. Like with the human blood, only the serum is needed and therefore is separated from the other ingredients. In the rabbits blood there are now specific antibodies against the human blood serum proteins. Mixing it with human blood again, an agglutination of the specific anti-bodys and the serum proteins takes place: a precipitation forms. This is the norm for the test as it is the maxium amount of preciptine that can form. Mixing the anti-human serum with the blood of other animals, a specific amount of antigen-complexes connect. The relationship is then determined by the amount of these compexes. The more of them form, the closer they are related. If you would do this test with a gorilla and a horse, you can see that a gorilla has over 80% accordance to the standardised sample and the horse just about 3%.
Amino acid sequence comparison
Proteins differ in the sequence of their amino-acids. The idea is, that the closer the compared amino-acids-sequences are, meaning they have similar amino acids at a certain position, the more related they are. Differences in the sequence are considered as mutations and happen over time. According to this, the more differences in the sequence are, the more time has passed in between the compared individuals and the less related they are.
Example:
Comparing cytochrome c, found in the inner membrane of a mitochondrion, from a human to any other breathing organisms (as it occurs in all of them) can be used. Thinking about an original cytochrome from which the alterations developed, after time passed it's structure stayed the same but also changed at some positions which can get compared and put into a matrix, calculating the distance (time passed) in a dendogram.
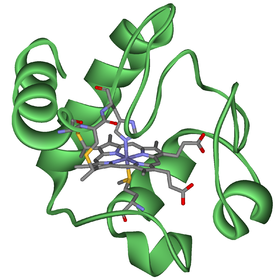
Structure of cytochrome c, an electron donator and acceptor in the respiratory chain
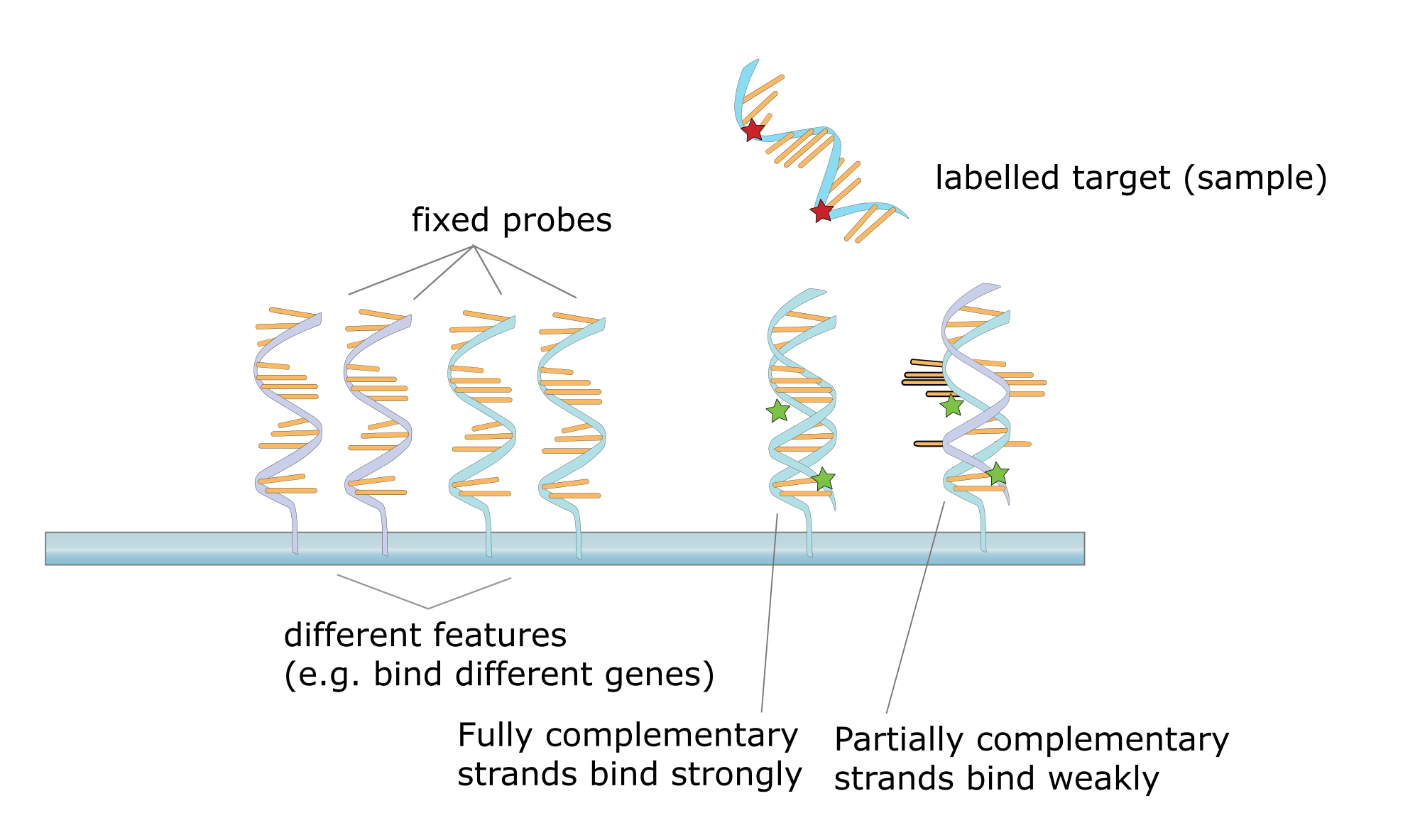
DNA-DNA-hybridisation
The genetic code Darwin didn't know about yet, makes such an analysis much more detailed. Breaking up the individuals DNA into two separate DNA single strands through high temperature, they are able to pair again after they cooled down again. This time they won't pair with their complementary strand, but with another single strand of another individual by reducing the temperature so they are able to form hydrogen-bridges again (renaturation). After this step, it is necessary to increase temperature again. The specific temperature for the break-up of the two strands is an indicator of how much hydrogen bridges were formed. A lower temperature means less hydrogen bridges, so the relationship is small too. Another way would have been to mark the individuals DNA and leave the DNA you want to compare to it unmarked. Pairing the two and "cutting of" the unbonded bases with an enzyme, the radioactivity is the indicator now.
Conclusion
These were some very basic test to determine relations between species. More accurate test require many information and not only one particular protein, avoiding simplifications and not only taking one gene as the representative for the whole organism's evolution. Other tests are used too and this was just a little overview over a very difficult field of biology.
SourceTexthttps://en.wikipedia.org/wiki/Darwinismhttp://www.biologie-schule.de/evolutionstheorie-darwin.php (translated)https://de.wikipedia.org/wiki/Serum-Präzipitin-Test (translated)http://ueevolution.blogspot.de/2014/08/methoden-4.html (translated)http://www.evolutionslehrbuch.info/index2.php?artikel=teil-5/kapitel-10-02.html (translated)http://www.spektrum.de/lexikon/biologie/cytochrome/16535 (translated)Pictureshttps://upload.wikimedia.org/wikipedia/commons/4/45/Charles_Darwin_1880.jpghttps://upload.wikimedia.org/wikipedia/commons/a/a2/Primate_cladogram.jpg/http://laboratoryinfo.com/wp-content/uploads/2015/05/Direct-coombs-test.jpghttps://upload.wikimedia.org/wikipedia/commons/thumb/0/07/Cytochrome_c.png/280px-Cytochrome_c.pnghttps://upload.wikimedia.org/wikipedia/commons/thumb/a/a8/NA_hybrid.svg/2000px-NA_hybrid.svg.png