Preface
In previous posts we have discussed:
Part 2: DNA Replication
Here we went into a bit more detail about the protein mechanisms by which a new copy of DNA gets created.
Part 3: Protein Expression
Here we learned about how information is extracted from DNA and is turned into the functional component of the cell known as a protein.
Part 4: DNA Damage
Here discussed a bit about what DNA damage means, and went through a few examples of types of damage. We also discussed what changes those damages can cause on newly generated DNA and the cell.
Today we will expand on the last posts discussion of DNA damage and learn about some of the ways those damages are repaired. These repair techniques employed by the cell involve a whole host of new proteins and some old favorites, and usually have consequences of their own. By that I mean the repair isn’t perfect, but the alternative is often cell death (and not even a cell wants to die!) Let us begin with the worst type of damage, the double strand break.
Double Strand Break Repair
In part 3 we discussed a type of DNA damage that results in the strand being split down the middle into two separate double stranded chunks. This is called a double strand break. There are two methods by which this type of damage gets repaired, non-homologous end joining and homologous recombination.
Non-homologous End Joining
This sort of repair is not directed by DNA sequence, a protein complex forms around two free ends of DNA (Including Ku and others Figure 1) and recruits an enzyme capable of linking them together called a DNA ligase (Figure 1, covered in part 2). The ligase seals each of the two strands back together making one whole piece of DNA. This sort of repair can lead to other issues if the ends of DNA that are put together are not the same two ends that broke apart (for instance if there are multiple breaks) resulting in things being randomly stitched back together.
Figure 1: Non Homologous End Joining
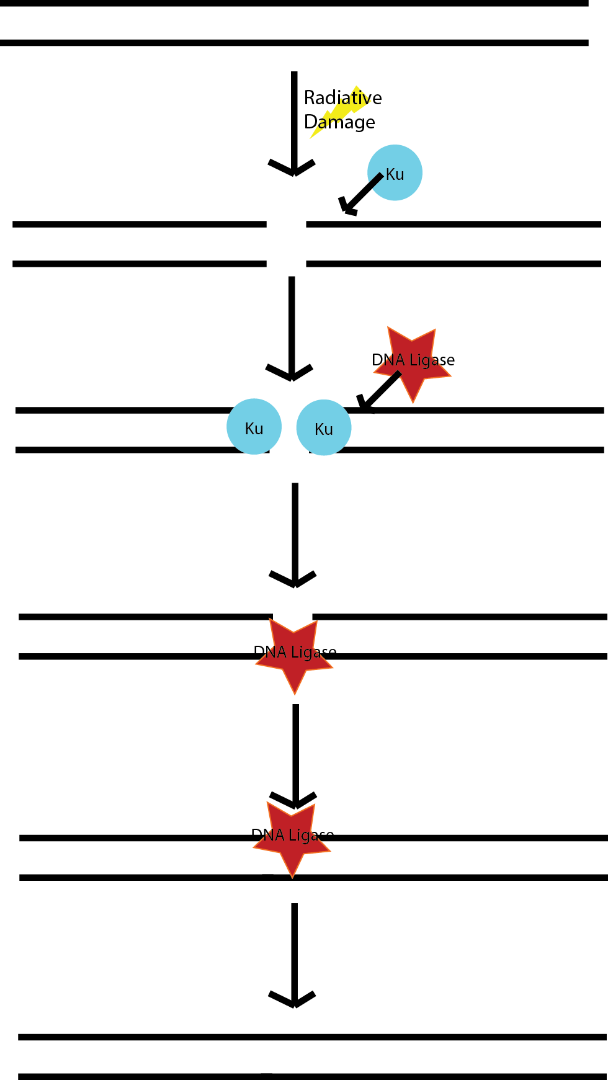
Source: Self
This can lead to a multitude of changes to any proteins that were encoded by that region of DNA and as we discussed previously, changing a proteins sequence is usually not good. Nevertheless from the perspective of the cell, an intact genome is better than a chopped up one, at least the intact one can be replicated.
Homologous Recombination
Homologous recombination is a repair pathway that occur during active cellular replication, and requires a separate piece of duplex DNA complementary to the one which has the double strand break. A complex of proteins called Rec proteins (I will simplify this complex and just call it Rec) binds to the broken DNA and unwinds it and chops back one of the strands from each side (Figure 2). This forms a single strand which then inserts itself into the second complementary duplex forming a “holiday junction” (the homology between the two pieces of DNA is why this is called homologous recombination, because the two pieces must be equal). The crossed strand is then extended (by a DNA polymerase, discussed in Part2) the holiday junction cut, and the two strands sealed by a DNA ligase (Enzyme and its function discussed in Part 2). However now the original sources of the DNA are now mixed (blue strands, tan strands and black strands have become mixed strands, Figure 2).
Figure 2: Homologous Recombination of Double Strand Breaks
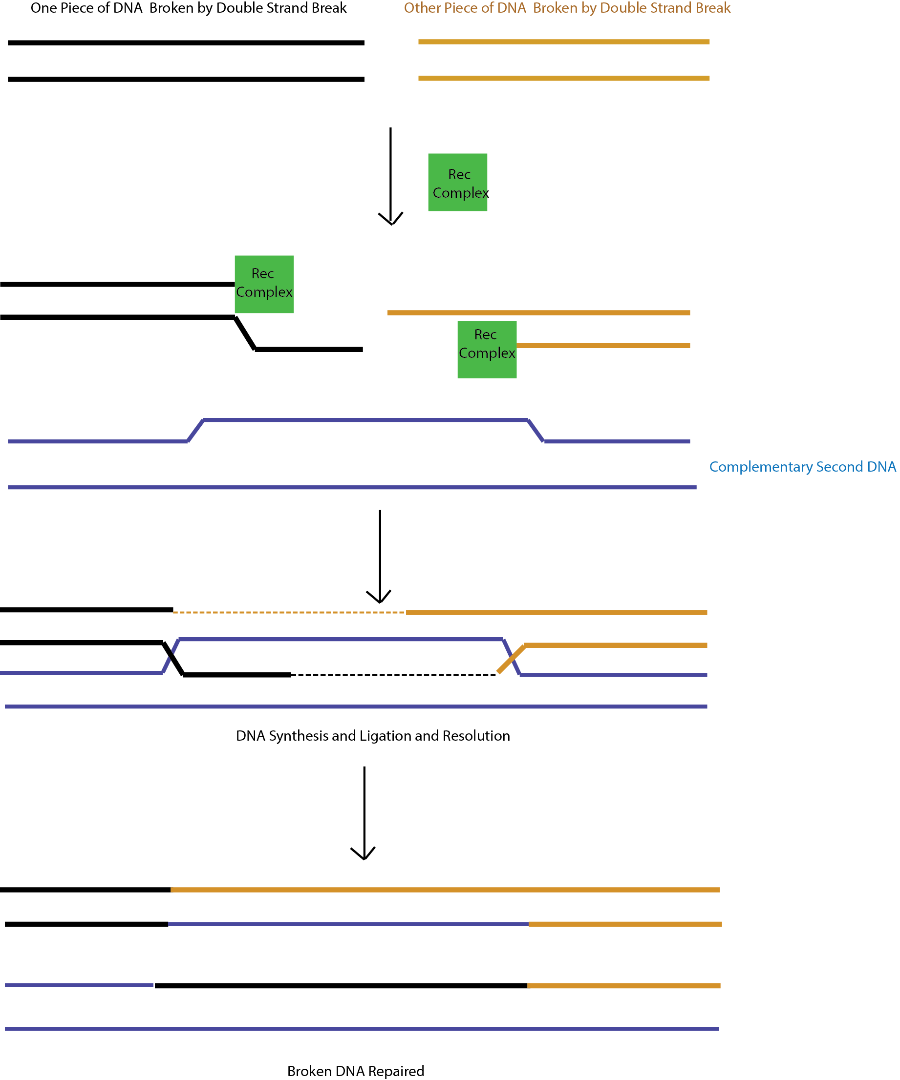
Source: Self
Repair of Other DNA Damage
Much like double strand breaks, the other forms of DNA damage each have their own specialized pathways to fix things.
For instance in Part 4 we discussed the formation of T-T dimers from UV light. When this happens there is a special enzyme called photolyase which is activated and can come in and cut out the damaged bases. Once they are cut out, and DNA polymerase can come in to save the day and replace the missing bases. Ligase seals the strands back up again and the damage is long forgotten. Processes like this one are slow however, so it works well, but only if the damage is at a minimum.
What happens when the cell is overwhelmed by DNA damage?
When a cell is overwhelmed by damaged bases and replication has come to a grinding halt, the SOS response is activated (SOS as in the international Morris code distress signal, and this response is exactly what you think it is, the cells “OH SHIT, I’M GONNA DIE” response). The whole purpose of this response is to cause the production of a special DNA polymerase(s). This/these polymerase(s) allow(s) for these damaged bases to be bypassed and replication to continue, but it comes at a huge cost. Mutations. The special damage induced polymerases don’t put in the correct bases opposite the mutations, they just throw in whatever they can. As we have discussed before, mutations can have a disastrous effect on the cell, but for the SOS response to be activated the damage must be so severe that the cells only alternative is death, so a few mutations are a fair price to pay.
Summary:
Cells have a variety of ways to deal with damage, but many come at a cost. Maintaining the genome is of utmost importance, and the consequences of poor maintenance are as we know, cell death and cancer. Each type of DNA damage often has its own unique set of solutions, and while most are effective they are not very fast. Too much damage can overwhelm the more effective repair tools, and lead to the SOS response, where the cell tries to fight for its life with special DNA polymerases.
Future Topics
What Is Cancer and What are Its Causes?
Genetic Modifications (eg. How are GMO Foods Made?)
Other Topics of Interest Suggested By YOU (still waiting for some :D )
Function and Preventative Care of Neurons (pending additional research on my part)
Please follow my blog and check out the previous entries in this series! Do not hesitate to DM me on steemit.chat where my username is also justtryme90. I would be happy to discuss my posts, science, answer any questions, or discuss anything else you would like!
