Part 1
Part 1.1-2
part 2
part 3
4.0 Sensitizers/derepressors
4.1 BAD
The pro-apoptotic BH-3 only protein BAD is regulated by posttranslational modification. In unstimulated cells BAD is unphosphorylated and associates with the anti-apoptotic BCL-2 repertoire. It has multiple phosphorylation sites and its pro-apoptotic function is inhibited by several kinases. Phosphorylation of BAD can result in reduced activity via cytoplasmic sequestration and inactivation by 14-3-3 proteins. 14-3-3 proteins remove phosphorylated BAD from the OMM and prevent BAD interaction with anti-apoptotic BCL-2 proteins. Pro-survival stimuli such as growth factors inactivate BAD via kinases such as Protein Kinase B (Akt) (Datta et al., 1997). Akt phosphorylates BAD at Serine-99, rendering it susceptible to OMM removal by 14-3-3 proteins, dissociating it from, for example BCL-xL. BCL-xL can then bind tBID promoting cell survival. Unphosphorylated BAD releases tBID from BCL-xL, allowing tBID to bind to and activate BAX (Lovell et al., 2008).
One of the critical phosphorylation sites on BAD is serine-75. This site is phosphorylated by protein kinase A (PKA), protein 90 ribosomal s6 kinase (p90RSK), and other proteins such as PAK1 and RAF-1 have been implicated to interact at this site (Jin et al., 2005: Polzien et al., 2009: Ye et al., 2011).
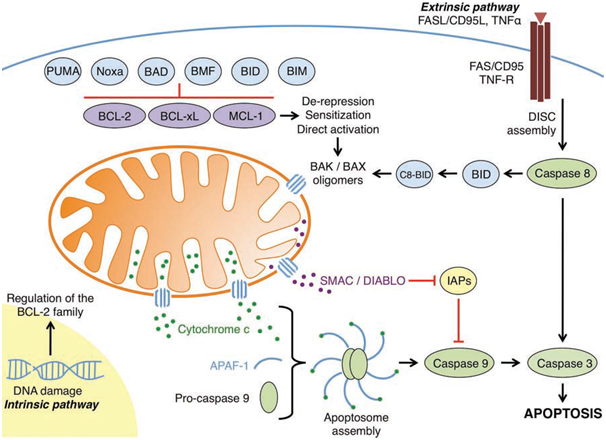
Figure 1: An overview of the intrinsic and extrinsic apoptotic pathways
(Elkholi et al., 2011).
BAD has been recently shown to also undergo posttranslational methylation by protein arginine methyltranferase-1 (PRMT1). PRMT1-mediated methylation of BAD occurs at arginine-94 and arginine-96 (Sakamaki et al., 2010). Methylation of these two arginine residues inhibits serine-99 phosphorylation of BAD by Akt. PRMT1 negatively inhibits Akt-mediated phosphorylation of BAD, and prevents the formation of BAD:14-3-3 complexes.
Several phosphatases also interact with BAD, such as protein phosphatase 1 α (PP1 α), PP2A, and PP2C during cytokine deprivation, dephosphorylating BAD and enhancing its pro-apoptotic activity.
BAD induces apoptosis via interaction with the anti-apoptotic BCL-2 family members. It forms heterodimers with anti-apoptotic proteins preventing inhibition of apoptotis. Another proapoptotic function of BAD is to free sequestered direct activators (for e.g. BIM) from anti-apoptotic proteins, allowing the direct activator to activate BAK and/or BAX, resulting in MOMP and cytochrome c release from the mitochondria. Although the pro-apoptotic activity of BAD has been shown to be regulated mainly by phosphorylation/dephosphorylation, and to a lesser extent methylation, other posttranslational modifications may also regulate BAD.
4.2 BIK
The BH3-only protein BCL-2-interacting killer (BIK) is tightly regulated by transcription factors, phosphorylation and cleavage. BIK is predominantly found in the ER and translocates to the OMM after apoptotic stimuli. Unlike most other BH3-only proteins BIK may have a well-defined structure (Hinds et al., 2007). BIK contains an exposed BH3 domain and a TMdomain. Due to its exposed BH3 domain BIK is thought to be constitutively active.
BIK is transcriptionally targeted by P53 after DNA damage, SMAD via TGF-β, and E2F1 during genotoxic stress (Real et al., 2006).
The apoptotic function of BIK is enhanced after phosphorylation. This phosphorylation of BIK occurs at threonine-33 and serine-35 by a casein II-like kinase (Verma et al., 2001). BIK has been shown to be rapidly degraded by the proteasome (Hur et al., 2006); phosphorylation of BIK may stabilise and protect it from degradation, much like the BH3-only protein BIM.
Rhomboid domain containing 1 (RHBDD1) regulates BIK-mediated apoptosis by proteolytic cleavage of BIK at its TM region (Wang et al., 2008).
BIK-mediated apoptosis is BAX-dependent. BIK interacts with anti-apoptotic proteins BCL-2 and BCL-xL. In unstimulated cells BAK is sequestered by BCL-2, BCL-xL and MCL-1, whereas BAX is predominantly sequestered by BCL-2, and BCL-xL. BIK is able to disrupt BAX:BCL-xL/BCL-2 complexes, freeing BAX to be activated and undergo OMM insertion/oligomerisation. Due to the inability of BIK to interact with MCL-1, it cannot free sequestered BAK. Another apoptotic function of BIK is inhibition of the anti-apoptotic ERK1/2 pathway. It interacts in a BH3-dependent manner and prevents nuclear translocalisation of ERK1/2, this inhibits cell survival. Other isoforms of BIK are thought to exist.
4.3 HRK
The BH-3 only protein harakiri (HRK) is regulated by transcription factors only. Enzyme-linked immosorbent assay (ELISA) and NMR data have shown HRK to be a largely unstructured protein (Sborgi et al., 2010) that contains a BH3 domain and a TM domain which inserts readily into intracellular organelle membranes such as the OMM (Barrera-Vilarmauet al., 2011). The transcription factor E2F1, upregulates HRK, as well as the other BH3-only proteins PUMA, NOXA, BIM and BIK (Hershko and Ginsberg, 2004). The transcription factor E2F1 highlights the control of one of the many synergistic mechanisms behind many of the BH3-only proteins apoptotic effects.
HRK exerts its pro-apoptotic activity through its BH3 domain by interacting with anti-apoptotic proteins Bcl-xL and BCL-2 (Inohara et al., 1997: Sborgi et al., 2010). HRK most likely drives apoptosis in a similar fashion to BIK by freeing sequestered BAX. ELISA and NMR data have shown that HRK can anchor to the OMM via binding to BCL-2 and BCL-xL independently of its TM domain (Barrera-Vilarmauet al., 2011). Membrane binding is an important aspect of the function of HRK, which backs up the embedded together model that suggests that most interactions within the BCL-2 family occur solely in the membrane.
Similar to tBID, HRK has been proposed to undergo myristolation at glycine-63 in the N-terminus of its TM domain, resulting in increased efficacy of OMM targeting.
HRK has recently been shown in vitro to interact with the BCL-2 multi-domain member Diva (Sborgi et al., 2010). More research is required between the interaction of HRK and Diva as this complex may not be functionally relevant to apoptosis.
4.4 Noxa
Noxa is a critical BH3-only protein that is regulated by transcriptional factors. Noxa is upregulated by p53 (Oda et al., 2000), and P73 after DNA damage. The p53-dependent transcription of this protein works in synergy with the p53-dependent transcription of PUMA. Though PUMA is responsible for most of the apoptotic activity in p53-mediated apoptosis, Noxa increases the efficacy of this process. Noxa binds with high affinity to MCL-1 and with lesser affinity to A1 (Chen et al., 2005). The BH3-only protein BAD works in synergy with Noxa, by binding to the rest of the anti-apoptotic repertoire. The binding of Noxa to MCL-1 results in proteasomal degradation of MCL-1 via unknown mechanisms.
Noxa requires its BH3 domain to induce degradation of MCL-1 and also for OMM localisation. This suggests that Noxa interacts with MCL-1 and A1 prior to associating with the OMM, which is in concordant with the embedded together model.
The mechanisms to which Noxa induces apoptotis are unclear and contradicting with other BH3-proteins. BIK-mediated apoptosis is BAX-dependent as it is unable to activate BAK due to its inability to interact with MCL-1; Noxa interacts with and causes degradation of MCL-1 but strangely appears to function chiefly in a BAX-dependent manner. This may be due to the binding affinity of each anti-apoptotic protein to BAX and BAK, or perhaps due to cooperation between Noxa and other proteins, driving it towards triggering BAX for oligomerisation. Does MCL-1 bind much more avidly to BAX? This would prevent BIK from acting in a BAX-dependent manner. The mechanisms of Noxa require elucidation to answer this question. Cooperation between BIK and Noxa has been shown to more efficiently induce BAX activation and cytochrome c release, than either protein acting alone, which confirms that both proteins act predominantly to activate BAX. Recently it has been shown that Noxa can interact with BCL-xL and that it is the only BH3-only member that frees sequestered tBID from MCL-1(Zhang L.,et al., 2011). This could mean that Noxa-mediated activation of BAX is a result of freeing tBID from MCL-1.
No data is yet available on whether the binding of Noxa to A1 leads to the degradation of A1. Noxa has been shown to affect mitochondrial integrity through multiple apoptotic pathways through distinct mechanisms (Zhang L.,et al., 2011). Also Noxa has been shown to directly activate BAK (Dai et al., 2011) highlighting the complex and important nature of this protein.
4.5 BMF
The BH3-only protein BCL-2 modifying factor (BMF) is another BH3-only protein that is intrinsically unstructured (Hinds et al., 2007). BMF is regulated by transcriptional factors, posttranslational modification, and sequestration. This protein seems to function by supporting the direct activator BIM. Both BIM and BMF are upregulated by SMAD proteins via TGF-β signalling (Pinon et al., 2008). BMF and BIM both through localisation to the cytoskeleton function by reacting to intracellular damage. BMF in healthy cells is sequestered to the cytoskeleton by interaction with the myosin V motor complex via DLC 2 (Puthalakath et al., 2001). After pro-apoptotic signalling BMF is released from the cytoskeleton where it translocates to the OMM and interacts with BCL-2, BCL-xL,and BCL-w freeing the direct activator BIM to activate BAX. BMF is released from the myosin V motor complex through phosphorylation by JNK (Lei and Davis, 2002).
Whereas phosphorylation by JNK upregulates BMF the kinase ERK2 downregulates BMF by directly phosphorylates it on serine-74 and serine-77 reducing its apoptotic ability by altering its interactions with DLC2, and the anti-apoptotic proteins (Shao and Aplin, 2012).
The mechanisms of BMF have not been fully elucidated, and this protein has been reported to have alternatively spliced isoforms which may implicate further modes of regulation. BMF is closely related to BIM and undergoes upregulation and downregulation via the same pathways, implicating these two proteins as functioning synergistically. Unlike its closest relative BIM, BMF has not been extensively studied, highlighting an area that needs more extensively researched.
@RiskDebonair
Adventure Capitalist of the Future
