Mutations in the DNA sequence of a protein can affect the morphology and functionality of that protein.
In the first part of this series I discussed about the exponentially decreasing costs of genome sequencing (full or partial), I introduced single nucleotide polymorphisms (SNPs) and I described a few SNPs associated with obesity.
For a refresher, please see post 1 here. For the sake of keeping things simple, I'm only going to remind about what SNPs are. According to the NIH Handbook of Genetics:
"Each SNP represents a difference in a single DNA building block, called a nucleotide. For example, a SNP may replace the nucleotide cytosine (C) with the nucleotide thymine (T) in a certain stretch of DNA."
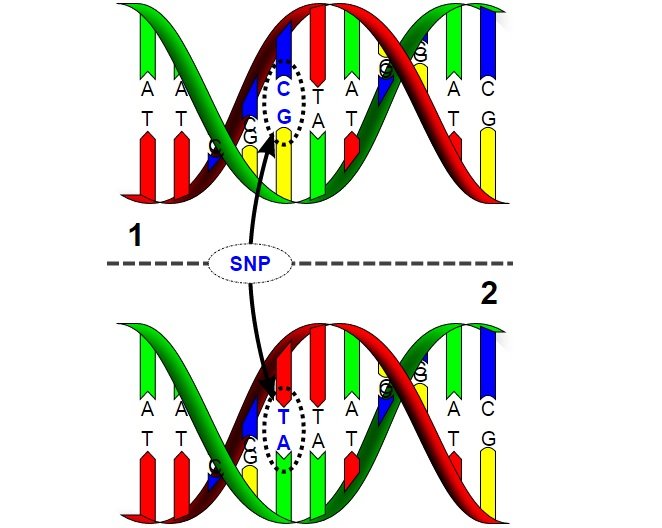
Each of us carry approximately 10 million SNPs, but many of them are not in parts of the genome to negatively impact physiology. Some of them are in non-coding regions. As it is pointed out by Genetics Learning Center of the University of Utah:
"SNPs and disease causing mutations are not the same."
In the first post I discussed about mutations (SNPs) in two apolipoproteins that have been consistently associated with obesity, the primary mechanism being a negative effect on lipid metabolism.
Here I'm going to talk about mutations in the FTO and MC4R genes.
Genetic Mutations (SNPs) associated with Obesity - Part 2
FTO - gene that codes for the Fat-Mass-and-Obesity-Associated protein
- strong association of FTO with BMI and the risks of developing obesity and T2D [source].
Important Single Nucleotide Polymorphisms of FTO (currently available):
1. rs1421085
The normal genotype for this position in the genome is TT, meaning that if you are TT at this location you have normal risk of developing obesity. The risk genotypes are CC and CT. If you have CC at this position you are at 1.7 times higher risk of developing obesity. If you have CT at this position you are at 1.3 times higher risk of developing obesity. One of the studies that found this association is here.
Another study from 2009 related to this is:
Read more about rs1421085.
2. rs1121980
The normal (average risk) genotype for this position is CC. People who are CT carry 1.67 times higher risk for developing obesity, while those who are TT carry 2.76 times higher risk for developing obesity - that is they are at almost triple the risk of becoming obese, which is alarming if you ask me.
The study that found these associations included ~1,000 Caucasians and it can be read here:
Read more about rs1121980.
3. rs16953002
The normal genotype for this position in the FTO gene is GG. People with low BMI who are GG have a slightly decreased risk of developing melanoma (skin cancer).
People who are AG or AA at this position carry 1.16 times, respectively 1.32 times higher risks of developing melanoma even though their BMI (body mass index) may be low.
The study to support these findings:
A variant in FTO shows association with melanoma risk not due to BMI.
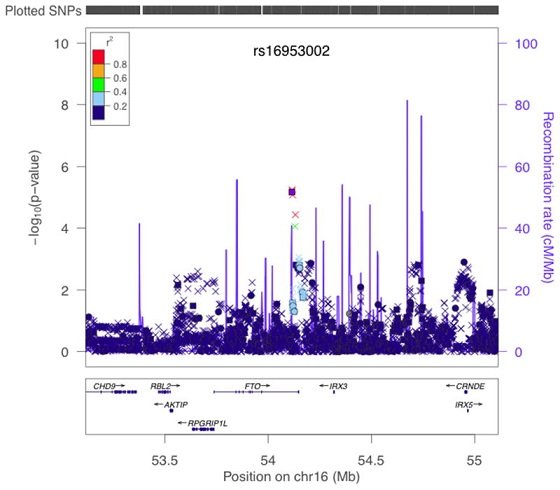
"Results of stratified trend tests of imputed data for association with melanoma in region around FTO in GenoMEL Phase 1 and 2 data combined. −log10p values for association between SNPs in the region of FTO and melanoma case-control status are shown adjusted for geographic region. Colour of points indicates degree of LD with rs16953002 (indicated by purple square). SNPs genotyped in all GenoMEL samples are plotted as circles, SNPs imputed in all samples as crosses and SNPs genotyped in some samples and imputed in others (as a result of chip differences) as squares. Positions of genes are given underneath the graph and estimated recombination rates also given by the blue line along the bottom, with scale on the right hand axis."[source].
Read more about rs16953002.
4. rs8050136
The normal genotype for this position in the FTO gene is CC (average risk). People who are AC or AA at this position are at 1.2 times, respectively 1.4 times higher risk of developing type 2 diabetes. To mention is that this finding is not consistent in all populations.
Read more about the studies and about rs8050136.
MC4R - gene that codes for Melanocortin 4 Receptor
According to the NIH:
"The protein encoded by this gene is a membrane-bound receptor and member of the melanocortin receptor family. The encoded protein interacts with adrenocorticotropic and MSH hormones and is mediated by G proteins. This is an intronless gene. Defects in this gene are a cause of autosomal dominant obesity."
Two important SNPs in the MC4R gene are:
1. rs2229616
The normal genotype for this position in the MC4R gene is GG. However, in this case, those carrying the normal allele may be disadvantaged. People who are AG or AA at this position at lower risk of developing diseases of the metabolic syndrome.
This is an example of beneficial/protective mutation. One of the studies to have found these associations included ~7,900 Europeans and can be read here:
Association of the MC4R V103I polymorphism with the metabolic syndrome: the KORA Study.
Read more about rs2229616.
2. rs17782313
The normal genotype for this position in the MC4R gene is TT (average risk). People who are CT or CC at this position are likely to be 0.22, respectively 0.44 BMI units higher - meaning they are prone to slightly increased body mass index.
The study to point this association included more than 60,000 adults and can be read here:
Common variants near MC4R are associated with fat mass, weight and risk of obesity.
Read more about rs17782313.
Ending Thoughts
This is a 'technical' post and if you are unfamiliar with genomics/sequencing related terminology, you may find some trouble reading/understanding it. However, if you decide to sequence your DNA for SNPs it may make much more sense and it can become helpful in understanding the genetic risks/advantages for developing/being protected from pathologic conditions.
A good intro into SNPs is provided in this 2-minute video explanation from 23andme:
You can use 23andme to find out about your SNPs. They currently look at 800k - 1 million locations in the genome. When you have the raw data from your genome sequencing, you can use services like Promethease to filter the most important data from your analysis. You wouldn't want to manually look for possible conditions associated with 1 million SNPs (many of them being unimportant/non-impacting to your physiology).
In my next post on genetic mutations I will continue the discussion on other obesity related SNPs.
To stay in touch with me, follow @cristi
Credits for Images: [David Hall CC BY 4.0 via Wikimedia Commons and [Iles et. al (2013) via NIH].
Cristi Vlad, Self-Experimenter and Author
