If you have high hopes that during your lifetime you'll be able to download your brain's content into a hard drive to avoid death or to become someone else's sex robot -Hey, I don't judge- I'll introduce you to the state of the art and the science of it.
The way the brain stores memories is theorized to be in biophysical and biochemical structures knows as engrams. The problem is where and how are they stored.
Form and function in biology have always been hand in hand. In the macro as morphophysiology (the anatomy and its role on function) and in the micro as structural biology (how the shape of molecules affects its function). Observation and manipulation for experimentation has always been the limiting rate for understanding this relationship.
For an organ like the brain, the observation of alterations inside the normal range is quite difficult. Outside of big and broad alterations, one can't find macro or micro differences in the anatomy.
The problem here is one of complexity. The diversity in cell types in the central nervous system is conspicuously big. This entails an enormous diversity of structure and function that is more specialized than in other tissues.
One can classify the cells in 2 big types. This as specific as saying a forest of trees (ambiguous), as there are many species of trees: 1011 Neurons (signaling cells) and 50x as many glia (the supporting tissue).
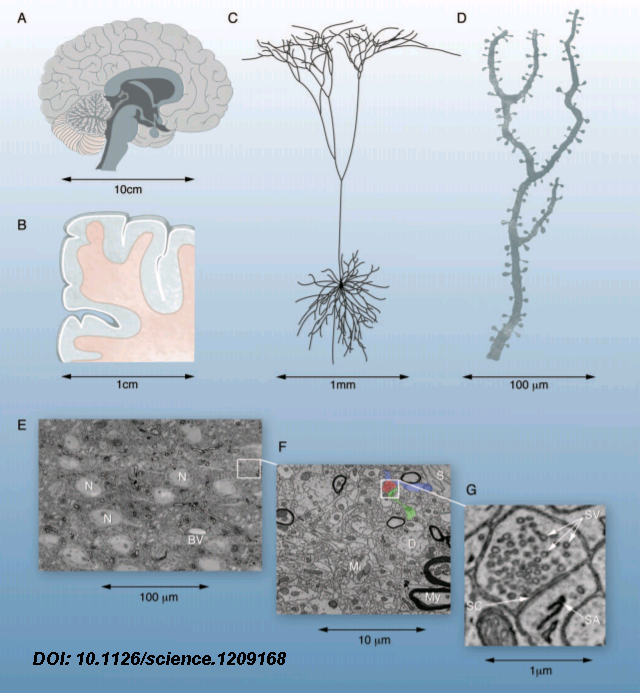
The scale of the brain goes from a dozen centimeters to a couple µm, 6 orders of magnitude. Studies of the brain are performed using imaging technology. The resolution of fMRI is of 1 cubic millimeter (1mm3). To give an idea an fMRI voxel is 1 trillion times larger than an electron microscopy voxel (100nm3) [1]
In other organs, the feedback between function and structure is more linear, from structure to function. In the case of the brain is more circular, like a loop. As some of the functions are shaped by experience rather than by genes.
In the human brain, in particular, there are critical periods of development. Processes like learning to walk, initiation of sexual maturity and independence take much longer in absolute and relative terms when compared to other animals. These developments are experience-dependent and are a form of outsourcing to the world for humans.
In order to map the network of connections one first needs to be able to differentiate a particular structure. Like with any problem of high complexity accurate representation of individual structures is key.
While techniques like the Golgi stain allow representation of the structure in cross-sectional cuts to derivate 3D information the result is a mess. That allows simple classification of layers without telling us anything about individual connections. The utility of the Golgi stain is that only 5% of the neurons stain, otherwise it would be too hard to discern anything.
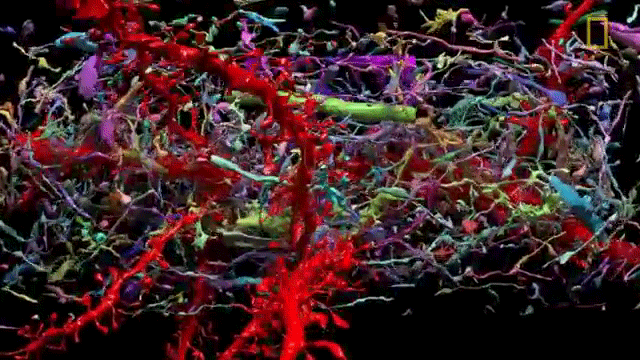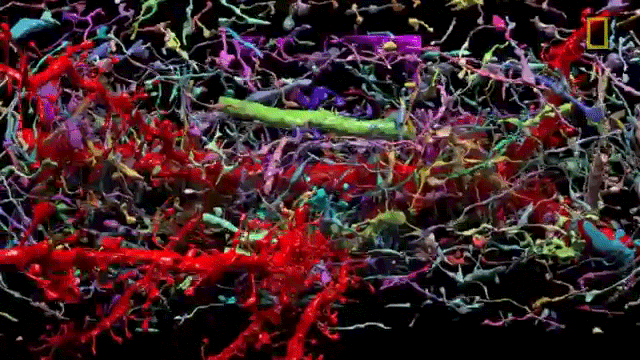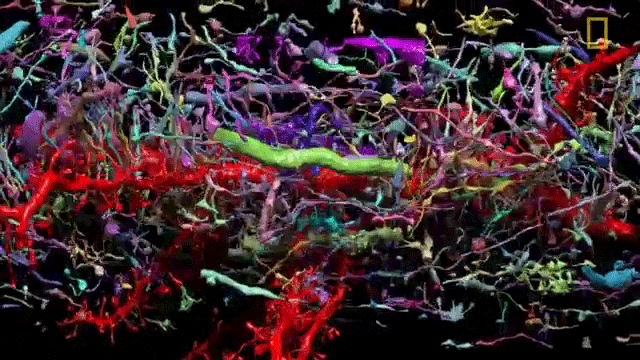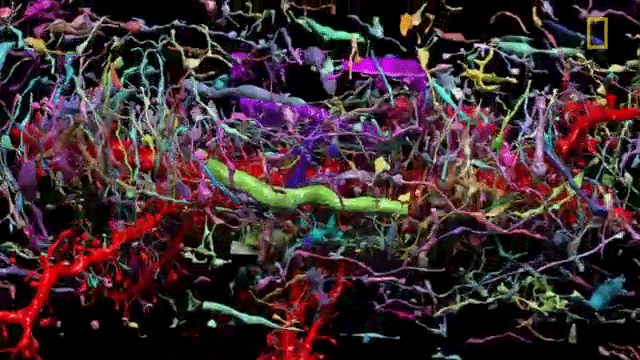
The problem of network visualization is one where computer hardware and informatics comes to aid. The complexity of the cables and the connection has been sorted by color labeling. [2]
We have adapted to represent and track highly complex information with color. A recent explosion in imaging has been possible thanks to the discovery of eGFP and derivated fluorescent proteins. Now antibodies allow amplification and more specificity by immunofluorescence. In the case of the central nervous system you can get most color combinations through 3 markers (anti-EGFP, anti-mOrange2, and anti-mKate2) the images produced are called the Brainbow[3].
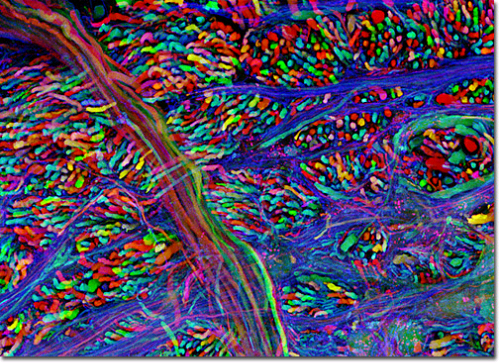
Using cre-recombinases that modify highly conserved promoter regions in the DNA of neurons, tandems of genes express fluorescent proteins randomly and homogeneously inside an individual neuron. Which makes it's color hues different to their neighbors.
This allows us to go deeper and map every synaptic vesicle at every synapsis. If there are close to 100 billion neurons and around 10.000 connections per neuron. That's roughly 1015 synaptic connections. Due to the storage of data and dynamic processing one could compare it to a 1.000 terabytes computer hard drive with a 1 trillion bit per second processor.
Aside from being great images as screensavers, the whole purpose of this is to create a map. Just like the Human genome project. The Human Connectome Project. A better name could be the synaptome but whatever.[4]
This approach is exhausting and fastidious (1.000 trillion connections). It has the disadvantages of only being usable in dead tissue. Fortunately, a new complementary technique appeared relatively recently. Diffusion weighted magnetic resonance imaging.
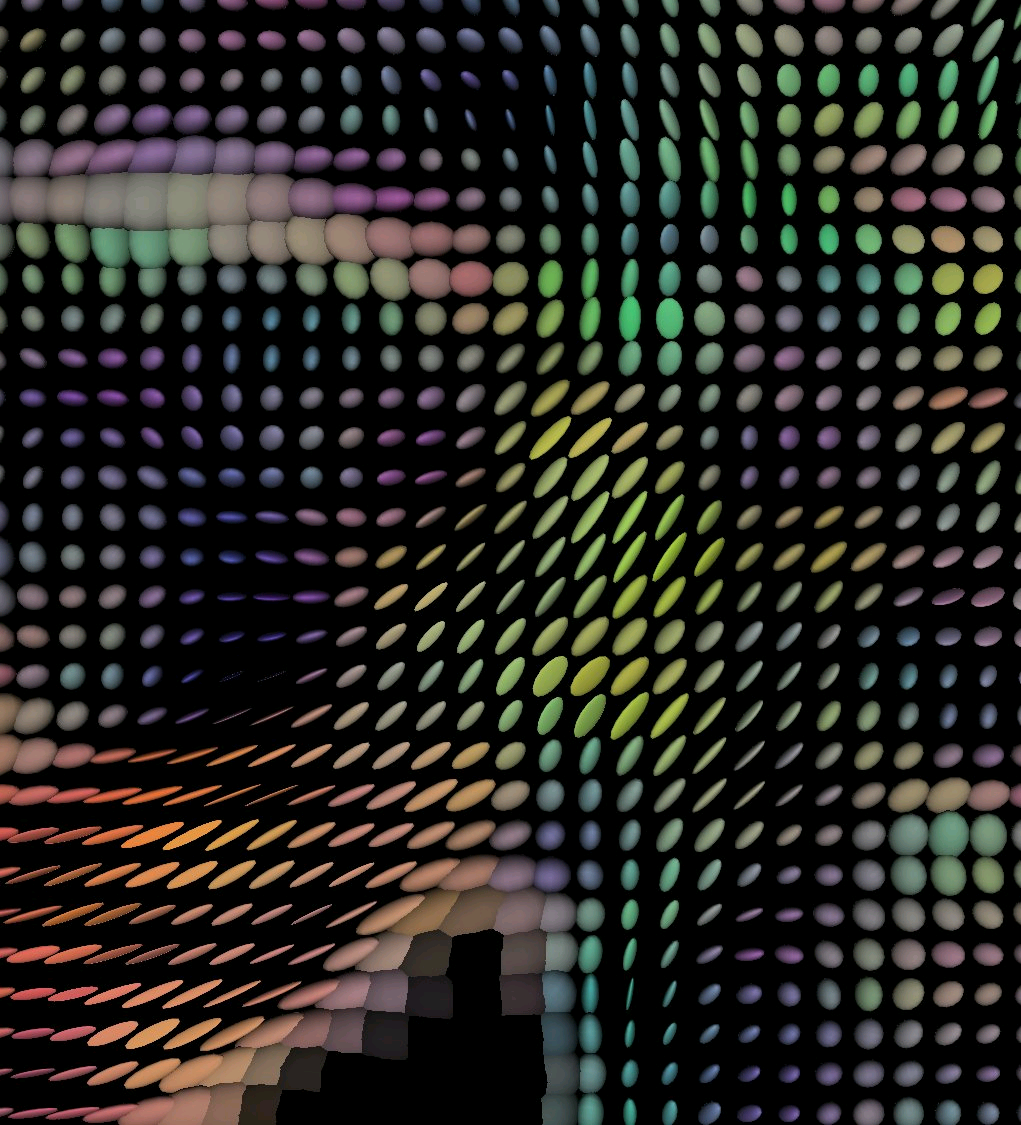 wiki commons Oblate spheroid voxels
wiki commons Oblate spheroid voxelsWater molecules inside tissues behave isotropically -equally in all directions- In the axons, due to the confined space, water behaves anisotropically. With more freedom to move in one axis, that of the direction of the axon.
Thanks to ever more powerful magnets and graphical processing power, it is now possible to create tractograms -imaging representations of the flow of water in the axons of the white mater- which allow live anatomical mapping by a particular variety of DW-MRI know as Diffusion tension magnetic resonance.
Here each voxel can be represented by an obloid shape that can toke one or more parameters that include a rate of difussion and a preferred direction of diffusion.
This type of visualization has great potential to map alterations in neuropsychiatric diseases that rely enormously on clinical criteria, despite some mathematical trade offs. [5]
So the idea is to simulate a whole brain. With all its functions. If once it starts running it behaves entirely as a real human brain the ethical dilemmas are interesting. Although that dilemma could be farther away than we would like to think.
In 2007 the BlueGene/L supercomputer of IBM simulated the equivalent of 1/2 a mouse brain for the equivalent of one second. As it was 1/10th of the speed. Current estimations of a full simulation of a human brain would require 1500x that speed and 1600 that capacity. The team of IBM expects to simulate the equivalent to a human cortex -880.000 processors- by 2019-2020.[6],[7]
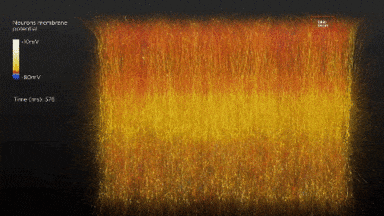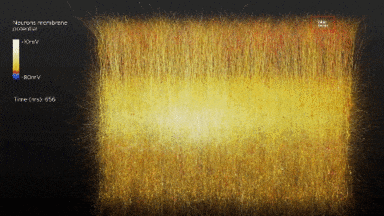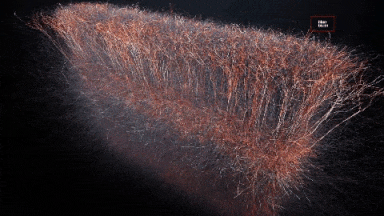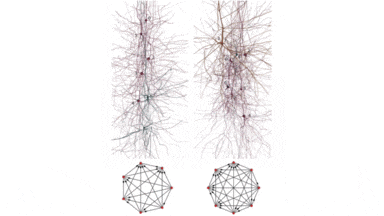
Another project BlueBrain by the Swiss government attempts to reverse engineer not only the processing power but to elucidate the mathematical relationships that govern the code of the brain. So far they have discovered several coding strategies of the central nervous system using algebraic topology.
An interesting fact linking structure and function is how the project discovered that in the neocortex, structures can achieve complexities of up to 11-dimenssions.[8] An interesting number of dimenssions for physicists or maybe just a spooky coincidence.
So far by today's standards, we can probably simulate the processing power of a really slow dog and the encoding power of a fruit fly.
What does the future await? Well, uploading your brain to a computer is not 100% impossible be happy. But the complexity of it makes it all look pretty distant. Also, we don't know if the connectome is sufficient or even necessary.
Sucks right?
References:
5 Tuch, D. S. (2004). Q-ball imaging. Magnetic Resonance in Medicine, 52(6), 1358–1372.
6 BBC news, technology. Mouse brain simulated on computer blueGene/L.
Images referenced, sourced or modified from google images, labeled for reuse